Toolkit-X: A better way to do DS&A problem lists
Introducing Toolkit-X: a free DS&A toolkit with curated BCtCI problems that can be used like a traditional problem list.
Introducing Toolkit-X: a free DS&A toolkit with curated BCtCI problems that can be used like a traditional problem list.
I am a computer scientist specialized in data structures and algorithms. Here is my resume.
Are you looking for a short bio? See the media kit.
I received a PhD as part of the CS Theory group at UCI. I was fortunate to be advised by David Eppstein and Michael Goodrich. Before that, I got a bachelor's degree in CS from UPC in my hometown, Barcelona.
My research spans computational geometry, greedy algorithms, graph data structures, computational biology, and recreational mathematics. My dissertation, New Applications of the Nearest-neighbor Chain Algorithm (pdf, blog post, advisor's blog post, defense slides) studies how to relax the "greedy choice" in certain greedy algorithms without affecting the final solution. This idea, paired with an algorithmic technique called nearest-neighbor chain, allows us to speed up some greedy algorithms (like the Multi-fragment algorithm for Euclidean TSP from O(n2) to O(n log n) (paper)).
See Research for more or download my academic CV.
After my PhD, I spent some time in industry as a senior SWE at Google. I worked on Google's internal software-defined WAN, optimizing the allocation of network bandwidth to Google's services.
I recently had the privilege to co-author Beyond Cracking the Coding Interview with Gayle Laakmann McDowell, Aline Lerner, and Mike Mroczka.
Not sure what's next yet, but lately I've been active on my blog.
My passion project is wallwars.net, an online board game.
Here's a little bit about my hobbies.

Click on a publication for a brief summary. See all publications for the complete list.
* N. Mamano, A. Efrat, D. Eppstein, D. Frishberg, M.T. Goodrich, S. Kobourov, P. Matias, V. Polishchuk. ISAAC'19
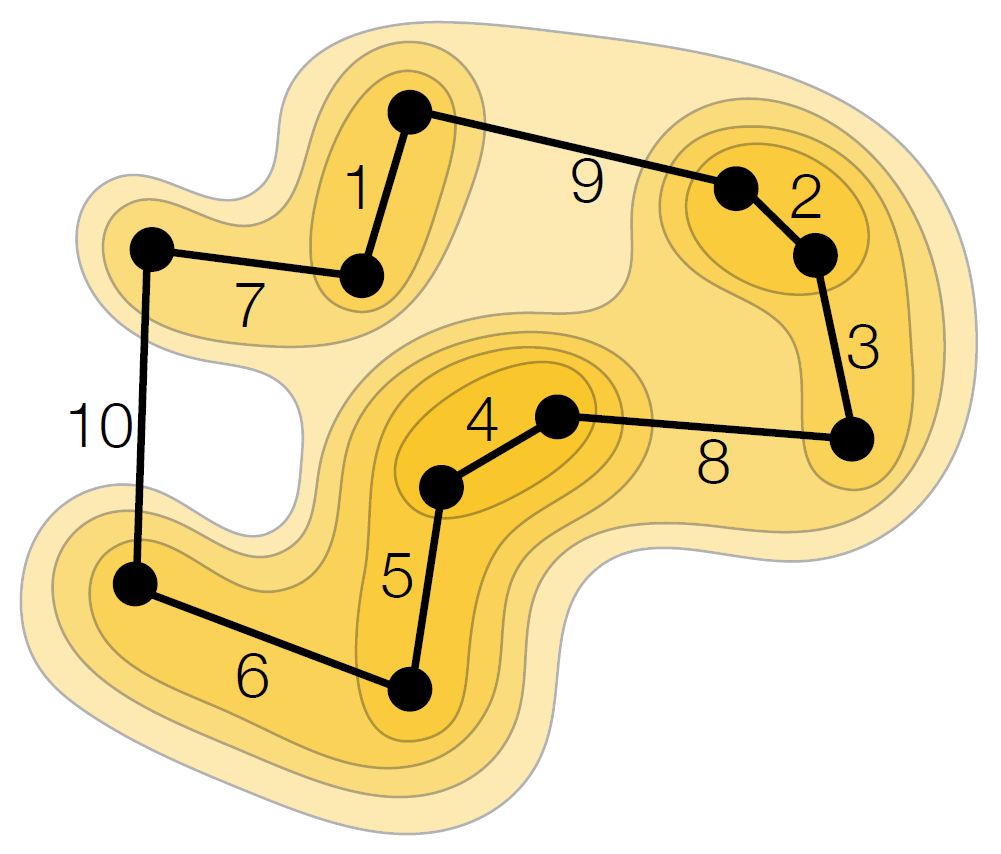
This paper contains some of the results from the thesis on speeding up greedy algorithms (read the thesis' summary above). For instance, we improve th...
* N. Mamano, W. Hayes. Bioinformatics: Oxford Journals, 2017
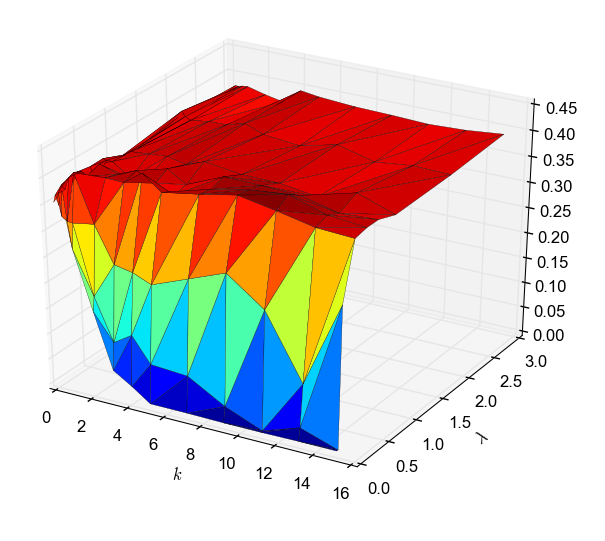
Protein-protein interaction networks are graphs where the nodes represent proteins and edges denote that two proteins can physically interact. Such gr...
Click on a project for a brief explanation. More projects on GitHub.